Near Duplicates Detection
Posted on February 19, 2023In my previous post I set up a tool to ease the download of open datasets into a JupyterLite environment, which is a neat tool to perform simplish data wrangling without local installation.
In this post we will put that tool to good use for one of the most common data cleaning utilities: near duplicate detection.

Why bother about near duplicates?
Near duplicates can be a sign of a poor schema implementation, especially when they appear in variables with finite domains (factors). For example, in the following addresses dataset:
| kind | name | number |
|---|---|---|
| road | Abbey | 3 |
| square | Level | 666 |
| drive | Mullholand | 1 |
| boulevard | Broken Dreams | 4 |
The “kind” variable could predictably take any of the following values:
- road
- square
- avenue
- drive
- boulevard
The problem is that this kind of data is too often modelled as an unconstrained string, which makes it error prone: ‘sqare’ is just as valid as ‘square’. This generates all kind of problems down the data analysis pipeline: what would happen if we analyze the frequency of each kind?
There are ways to ensure that the variable “kind” can only take one of those values, depending on the underlying data infrastructure:
- In relational databases one could use domain types , data validation triggers, or plain old dictonary tables with 1:n relationships.
- Non-relational DBs may have other ways to ensure schema conformance, e.g. through JSON schema or XML schema.
- The fallback option is to guarantee this “by construction” via application validation, (eg. using drop-downs in the UI), although this is a weaker solution since it incurs in unnecessary coupling… and thing can go sideways anyway, so in this scenario you should consider performing periodic schema validation tests on the data.
Notice that all of these solutions require a priori knowledge of the domain.
But what happens when we are faced with an (underdocumented) dataset and asked to use it as a source for analysis? Or when we are asked to derive these rules a posteriori eg. to improve a legacy database? Well, without knowledge of the domain, it is just not possible to decide wether two similar values are both correct (and just happen to be spelled similarly) or a misspelling. The best thing we can do is to detect which values are indeed similar and raise a flag.
This is when the techniques explained in this blog post come handy.
The algorithm
For the sake of simplicity, in this blog post we will assume our data is small enough so that a quadratic algorithm is acceptable (for the real thing, see the references at the end). Beware that, in modern hardware, this simple case can take you farther than you would initially expect. My advise is to always use the simplest solution that gets the job done. It usually pays off in both development time and incidental complexity (reliance on external dependencies, etc…).
There are two main metrics regarding similarity. The first one, restricted to strings, is the Levenshtein (aka edit) distance and represents the number of edits needed to go from one string to another. This metric is hard to scale in general, since it requires pairwise comparison.
The other one is both more general and more scalable. It involves generating n-gram sets and then comparing them using a set-similarity measure.
N-gram sets
For each string, we can associate a set of n-grams that can be derived from it. N-grams (sometimes called shingles) are just substrings of length n. A typical case is n=3, which generates what is known as trigrams. For example, the trigram set for the string "algorithm" would be ['alg', 'lgo', 'gor', 'ori', 'rit', 'ith', 'thm'].
Jaccard Index
Once we have the n-gram set for a string, we can use a general metric for set similarity. A popular one is the Jaccard Index. Which is defined as the ratio between the cardinality of intersection over the cardinality of the union of any two sets.
\[J(A,B) = \frac{|A \bigcap B|}{|A \bigcup B|}\]
Note that this index will range from 0, for disjoint sets, to 1, for exactly equal sets.
If we were to scale…
The advantadge of using n-gram sets is that we can use similarity-preserving summaries of those sets (eg. via minhashing) which, combined with locality sensitive hashing to efficiently compare pairs of sets provides a massively scalable solution. In this post we will just assume that the size of our data is small enought so that we do not need to scale.
The Code
All the above can be implemented in the following utility function, which will take an iterable of strings and the minimum jaccard similarity and max levenshtein distance to consider a pair a candidate for duplicity. It will return a pandas dataframe with the pair indices, their values, and their mutual Levenshtein and Jaccard distances. We will use the Natural Languate Toolkit for the implementation of those distances.
Bear in mind that, in a real use case, we would very likely apply some normalization before testing for near duplicates (eg. to account for spaces and/or differences in upper/lowercase versions).
def near_duplicates(factors, min_jaccard: float, max_levenshtein: int):
trigrams = [ set(''.join(g) for g in nltk.ngrams(f, 3)) for f in factors ]
jaccard = dict()
levenshtein = dict()
for i in range(len(factors)):
for j in range(i+1, len(factors)):
denom = float(len(trigrams[i] | trigrams[j]))
if denom > 0:
jaccard[(i,j)] = float(len(trigrams[i] & trigrams[j])) / denom
else:
jaccard[(i,j)] = np.NaN
levenshtein[(i,j)] = nltk.edit_distance(factors[i], factors[j])
acum = []
for (i,j),v in jaccard.items():
if v >= min_jaccard and levenshtein[(i,j)] <= max_levenshtein:
acum.append([i,j,factors[i], factors[j], jaccard[(i,j)], levenshtein[(i,j)]])
return pd.DataFrame(acum, columns=['i', 'j', 'factor_i', 'factor_j', 'jaccard_ij', 'levenshtein_ij'])We can extend the above functions to explore a set of columns in a pandas data frame with the following code:
def df_dups(df, cols=None, except_cols=[], min_jaccard=0.3, max_levenshtein=4):
acum = []
if cols is None:
cols = df.columns
if isinstance(min_jaccard, numbers.Number):
mj = defaultdict(lambda : min_jaccard)
else:
mj = min_jaccard
if isinstance(max_levenshtein, numbers.Number):
ml = defaultdict(lambda: max_levenshtein)
else:
ml = max_levenshtein
for c in cols:
if c in except_cols or not is_string_dtype(df[c]):
continue
factors = df[c].factorize()[1]
col_dups = near_duplicates(factors, mj[c], ml[c])
col_dups['col'] = c
acum.append(col_dups)
return pd.concat(acum)If we apply the above code to the open dataset from the last blog post
The column names are in Catalan since the dataset comes from the Barcelona Council Open Data Hub, and stand for the contractor, the service descripction, and the type of service.
We get the following results:
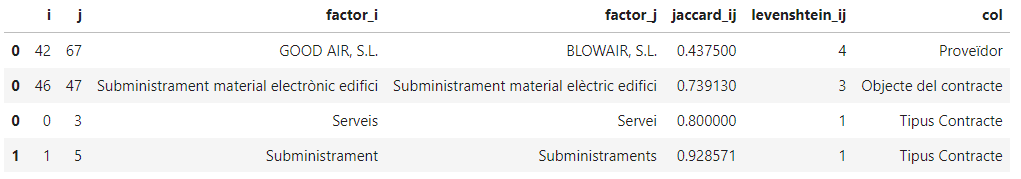
Notice that the first two are actually valid, despite being similar (two companies with similar names and electric vs electronic supplies), while the last two seem to be a case of not controlling the variable domain properly (singular/plural entries). We should definitely decide for a canonical value (singular/plural) for the column “Tipus Contracte” before we compute any aggregation for those columns.
Conclusions
We can use the above functions as helpers prior to performing some analysis on datasets where domain rules have not been previously enforced. They are compatible with JupyterLite, so no need to install anything for the test. For convenience, you can find a working notebook in this gist.
References
- Mining Of Massive Datasets - An absolute classic book. Chapter 3, in particular, describes a scalable improvement on the technique described in this blog post.